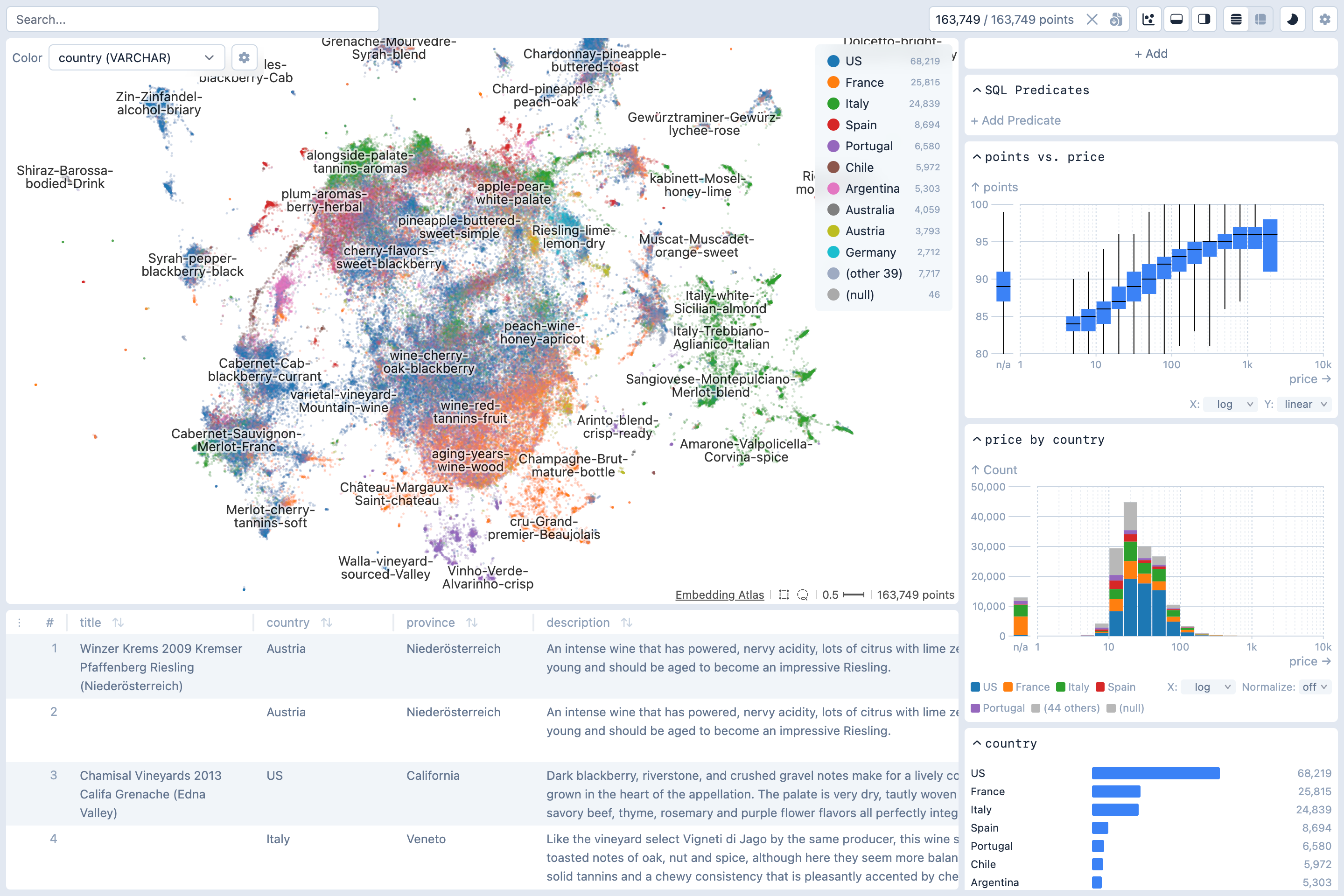
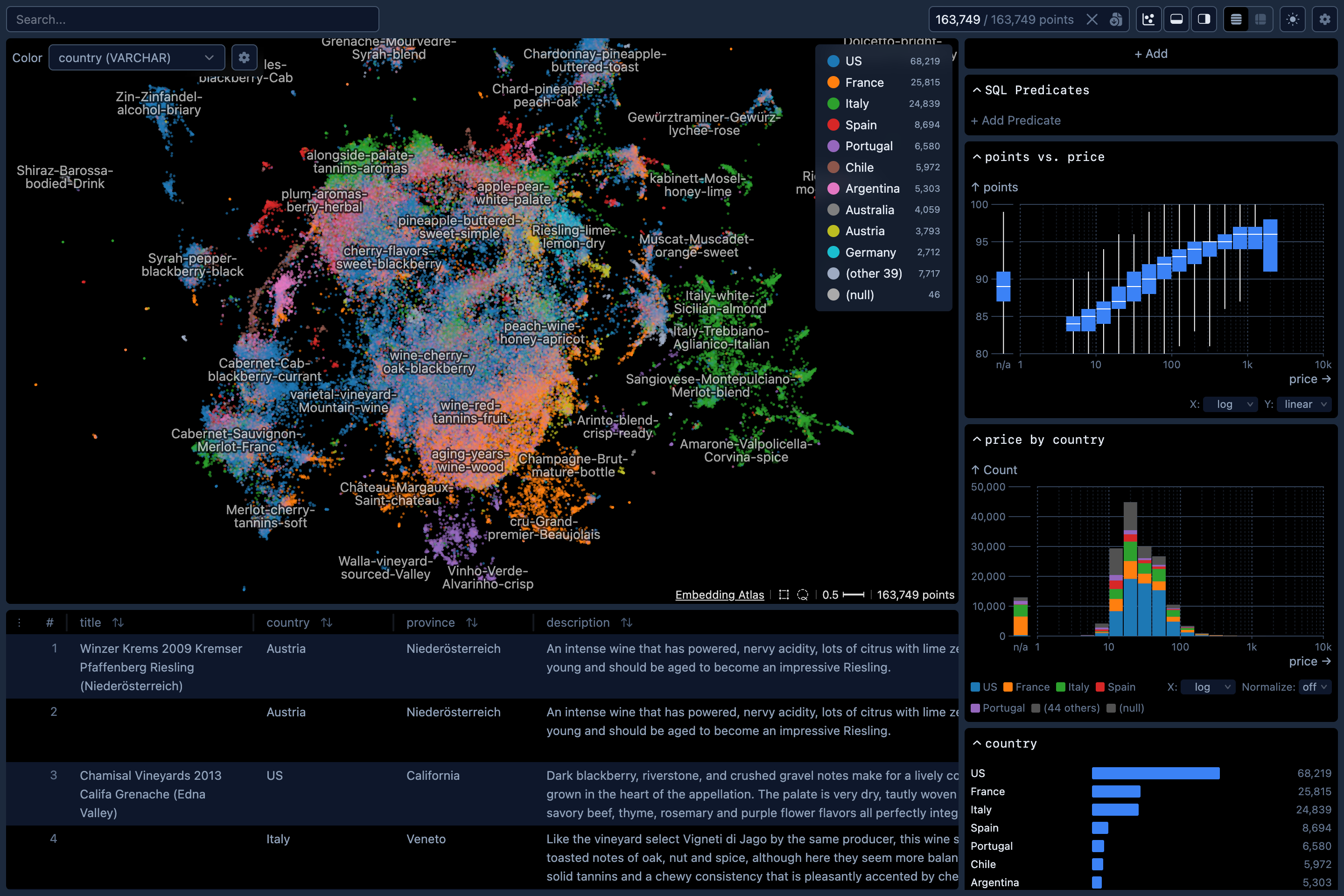
Command Line Utility
The Python package contains a command-line utility for you to quickly explore large text datasets with metadata.
Installation
pip install embedding-atlasand then launch the command line tool:
embedding-atlas [OPTIONS] INPUTS...TIP
To avoid package installation issues, we recommend using the uv package manager to install Embedding Atlas and its dependencies. uv allows you to launch the command line tool with a single command:
uvx embedding-atlasOn Windows, you may install the package on either the Windows Subsystem for Linux (WSL) or directly on Windows. To use NVIDIA GPUs, you'll need to install a PyTorch version that supports CUDA, see here for more details.
Loading Data
You can load your data in two ways: locally or from Hugging Face.
Loading Local Data
To get started with your own data, run:
embedding-atlas path_to_dataset.parquetLoading Hugging Face Data
You can instead load datasets from Hugging Face:
embedding-atlas huggingface_org/dataset_nameVisualizing Embeddings
The script will use SentenceTransformers to compute embedding vectors for the specified column containing the text or image data. You may use the --model option to specify an embedding model. If not specified, a default model will be used. The current defaults are all-MiniLM-L6-v2 for text and google/vit-base-patch16-384 for images, but these are subject to change in future releases.
After embedding vectors are computed, the script will then project the high-dimensional vectors to 2D with UMAP.
TIP
Optionally, if you know what column your text data is in beforehand, you can specify which column to use with the --text flag, for example:
embedding-atlas path_to_dataset.parquet --text text_columnSimilarly, you may supply the --image flag for image data, or the --vector flag for pre-computed embedding vectors.
If you've already pre-computed the embedding projection (e.g., by running your own embedding model and projecting them with UMAP), you may store them as two columns such as projection_x and projection_y, and pass them into embedding-atlas with the --x and --y flags:
embedding-atlas path_to_dataset.parquet --x projection_x --y projection_yYou may also pass in the --neighbors flag to specify the column name for pre-computed nearest neighbors. The neighbors column should have values in the following format: {"ids": [id1, id2, ...], "distances": [d1, d2, ...]}. The IDs should be zero-based row indices. If this column is specified, you'll be able to see nearest neighbors for a selected point in the tool.
Once this script completes, it will print out a URL like http://localhost:5055/. Open the URL in a web browser to view the embedding.
Reproducibility
For reproducible embedding visualizations, we recommend pre-computing both the embedding vectors and their UMAP projections, and storing them with your dataset. This ensures consistency since the default embedding model may change over time, floating-point precision may vary across different devices, and UMAP introduces randomness through both its default random initialization and its use of parallelism (see here).
The embedding_atlas package provides utility functions to compute the embedding projections:
from embedding_atlas.projection import compute_text_projection
compute_text_projection(df, text="text_column",
x="projection_x", y="projection_y", neighbors="neighbors"
)MCP Support
The command line utility supports Model Context Protocol (MCP). You can enable it with the --mcp flag. When running, it exposes an MCP server that allows AI agents to query the data schema, run SQL queries, create and modify charts, adjust the layout, and capture screenshots.
Usage
Usage: embedding-atlas [OPTIONS] INPUTS...Command Line Options
--text text
Column containing text data.
--image text
Column containing image data.
--vector text
Column containing pre-computed vector embeddings.
--split text
Dataset split name(s) to load from Hugging Face datasets. Can be specified multiple times for multiple splits.
--enable-projection / --disable-projection boolean
Compute embedding projections from text/image/vector data. If disabled without pre-computed projections, the embedding view will be unavailable.
--model text
Model name for generating embeddings (e.g., 'all-MiniLM-L6-v2').
--trust-remote-code boolean
Allow execution of remote code when loading models from Hugging Face Hub.
--batch-size integer
Batch size for processing embeddings (default: 32 for text, 16 for images). Larger values use more memory but may be faster.
--text-projector choice
Embedding provider: 'sentence_transformers' (local) or 'litellm' (API-based).
--api-key text
API key for litellm embedding provider.
--api-base text
API endpoint for litellm embedding provider.
--dimensions integer
Number of dimensions for output embeddings (litellm only, supported by OpenAI text-embedding-3+).
--sync boolean
Process embeddings synchronously (litellm only). Use for local servers like Ollama to avoid memory issues.
--x text
Column containing pre-computed X coordinates for the embedding view.
--y text
Column containing pre-computed Y coordinates for the embedding view.
--neighbors text
Column containing pre-computed nearest neighbors in format: {"ids": [n1, n2, ...], "distances": [d1, d2, ...]}. IDs should be zero-based row indices.
--query text
Use the result of the given SQL query as input data. In the query, you may refer to the original data as 'data'.
--sample integer
Number of random samples to draw from the dataset. Useful for large datasets. If query is specified, sampling applies after the query.
--umap-n-neighbors integer
Number of neighbors to consider for UMAP dimensionality reduction (default: 15).
--umap-min-dist float
The min_dist parameter for UMAP.
--umap-metric text
Distance metric for UMAP computation (default: 'cosine').
--umap-random-state integer
Random seed for reproducible UMAP results.
--duckdb text
DuckDB connection mode: 'wasm' (run in browser), 'server' (run on this server), or URI (e.g., 'ws://localhost:3000').
--host text
Host address for the web server (default: localhost).
--port integer
Port number for the web server (default: 5055).
--auto-port / --no-auto-port boolean
Automatically find an available port if the specified port is in use.
--cors text
Allow cross-origin requests. Use --cors to allow all origins, or --cors http://example.com for a specific domain (or a comma-separated list of domains).
--static text
Custom path to frontend static files directory.
--export-application text
Export the visualization as a standalone web application to the specified ZIP file and exit.
--with text
Import the given Python module before loading data. For example, you can use this to import fsspec filesystems. Can be specified multiple times to import multiple modules.
--point-size float
Size of points in the embedding view (default: automatically calculated based on density).
--stop-words text
Path to a file containing stop words to exclude from the text embedding. The file should be a table with column 'word'
--labels text
Path to a file containing labels for the embedding view. The file should be a table with columns 'x', 'y', 'text', and optionally 'level' and 'priority'
--mcp / --no-mcp boolean
Enable MCP (Model Context Protocol) server endpoints for external tool integration.
--version boolean
Show the version and exit.